
- #Jmol first glance mac os
- #Jmol first glance software
- #Jmol first glance series
- #Jmol first glance free
laboració.Įste correo electrónico y sus anexos pueden contener información confidencial o legalmente protegida y está exclusivamente dirigido a la persona o entidad destinataria.Si heu rebut aquest correu electrònic per error, us preguem que n’informeu al remitent i que elimineu del sistema el missatge i el material annex que pugui contenir. Si no sou el destinatari final o la persona encarregada de rebre’l, no esteu autoritzat a llegir-lo, retenir-lo, modificar-lo, distribuir-lo, copiar-lo ni a revelar-ne el contingut. In my opinion (I am using chrome) this is a not a trivial problem, especially using big proteinsĪquest correu electrònic i els annexos poden contenir informació confidencial o protegida legalment i està adreçat exclusivament a la persona o entitat destinatària. Even with the popup tool and extending the molecule through the whole screen the rotation is excellent.
#Jmol first glance software
#Jmol first glance series
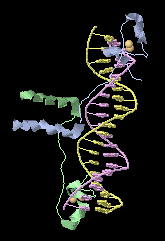
In order to be as easy as possible to use, FirstGlance in Jmol does not provide tools to customize the molecular view, but rather offers a series of "canned" views that reveal the major structural features of the molecule, such as composition (protein, DNA, RNA, ligand and solvent) secondary structure, amino and carboxy termini, hydrophobic and polar surfaces, surface charges, salt bridges and cation-pi interactions. Tooltips are used extensively, and explanatory help, with color keys, appears automatically for each view. It operates from menus and forms - no familiarity with the Jmol command scripting language is required.
#Jmol first glance mac os
It works in the most popular web browsers, and MS Windows, Apple Mac OS X, and linux.
#Jmol first glance free
It uses free open-source Jmol, as does Proteopedia. FirstGlance in Jmol version 4.0 was released August 15, 2022, with new capability to automatically construct large assemblies such as virus capsids.įirstGlance in Jmol ( ) is a free, open-source, macromolecular visualization software package that operates on-line in a web browser. When you go directly to FirstGlance in Jmol, you can enter any PDB code, or upload a PDB file from your computer, such as a prediction by AlphaFold or a homology model.įirstGlance in Jmol provides an easy interface to make a Powerpoint-ready animation from any of its molecular views. Every molecule in Proteopedia can be explored in FirstGlance in Jmol, using the link beneath the molecule (on pages titled with a PDB code, such as 1d66).


 0 kommentar(er)
0 kommentar(er)
